Jaw Volume Calculation
Update: Code added on github. Click here to view.
This project was done as part of a research on volume expansion of a jaw after using arch expansion treatment. Worked with Doctor Pooja Radhakrishnan to identify the volume of a jaw from a 3D medical scan.
Stage 0: The initial plan was to manually mark edge points of jaw and calculate volume of the trapezoidal shape formed. However, this has large scope for manual error, and so other options had to be considered.
Stage 1: The first step in order to find the volume using MATLAB, was to separate the bone from the soft tissue in the CBCT scan. In this scan, the intensity value for each voxel is proportional to the density of the area being scanned. The bone has a higher intensity than soft tissue in the image. Therefore, I separated the bone by deciding a fixed intensity threshold. I later realised that a fixed threshold was not very accurate, and so changed it then.
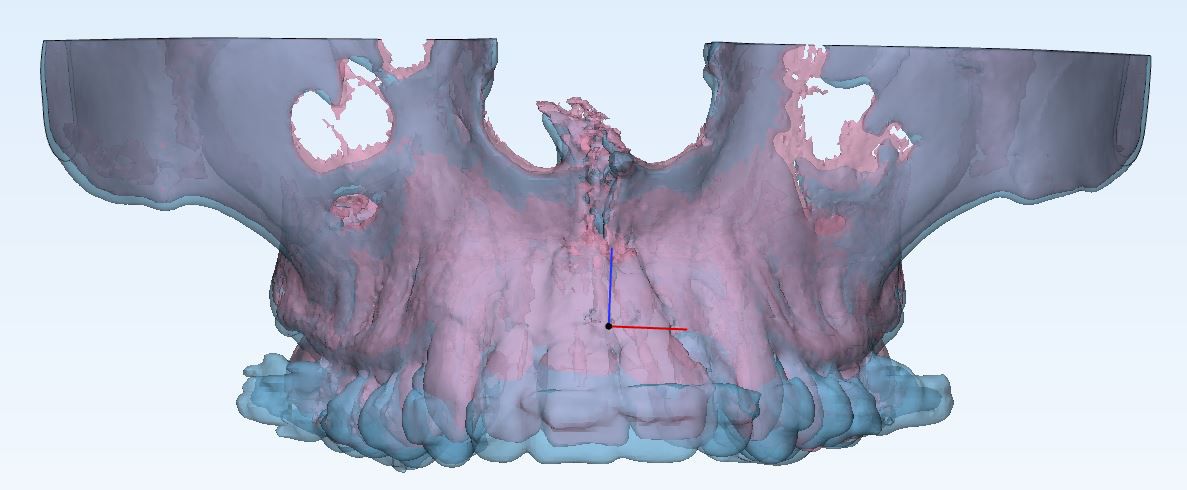
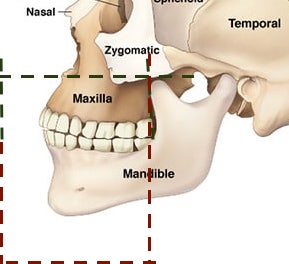
Stage 2: The next step was to separate the jaw from the rest of the image. I tried multiple approaches; using segmentation algorithms, using the teeth as reference for the jaws, and slicing the image at fixed points. Uassing betweeltimately, the last one showed best results and so we used that. the upper jaw (Maxilla) was bounded on top by the nasal canal, and at the back by the bone of the lower jaw. The lower jaw (Mandible) was bounded at the back at the point where the upper end of the lower jaw began. The maxilla and mandible were separate by a plane passing between the jaws.
Stage 3: Once these steps were done, the volume of the remaining black and white image was to be found. The simplistic approach implemented was to find the number voxels set to 1. Without an idea of what value could be expected, it was unclear how accurate the results were. Once the pre and post scans were both available, we noticed that the volume difference calculated was erratic; ranging from 5 mm^2 to 2000 mm^2. A few values were also negative, proving that we needed more accurate volume calculation.
Stage 4: The first thing I noticed is that the pre and post scans were misaligned. And so I used registration to align the two images before the thresholding and segmenting steps conducted on each image with the same threshold values and cropping planes. While this improved the accuracy somewhat, we still received some negative and erratic values.
Stage 6: At this stage, I felt the need of expert guidance to move forward. I did feel that the thresholding method could be improved, as each image had different histograms. I contacted Professor Venkatesan Rajinikanth and informed him of our situation. He not only provided an improved thresholding algorithm, which was already helpful, but also suggested I use the Gray Level Co-Occurrence Matrix (GLCM) method that extracts details about texture, to find the volume of the bone.
Stage 7: An unexpected suggestion, I did my research on the topic and found that it might work. I am currently working on implementing this solution, and am confident that we will get accurate results using it. In the mean time, the results are also being calculated with the help of Materialise Mimics and Autodesk Meshmixer software.